CODE 1D DPFT
- Date:
8 Oct 2019
Note
The .rst file for this page is generated using Jupyter Notebook, from the .ipynb file: 1D DPFT In Python
Thomas Fermi Kinetic Energy
Where \(\alpha \equiv {\pi^2\hbar^2\over 2mg^2}\)
And \(g\) is spin multiplicity. For spin-polarized fermi gas (For example electron gas with all the electrons spin-up), \(g=1\) from Pauli principle
Without Interaction
is a Functional of \(\mu-V_{\text{ext}}\)
Where \(\mu\) is the chemical potential
The Interaction
Where \(w\) is strength of interaction
Then we have
Self Consistent Loop
What is the idea?
We will find \(n\) with interaction
However, we will NOT use the expression \(n_{\text{int}}\) above
Instead, we use the expression \(n_{\text{nonInt}}\):
Specifically we will use \(n=\sqrt{\alpha^{-1}[\mu-V]_+}\)
We initialize \(V\) with \(V_{\text{ext}}\)
Then we find the chemical potential \(\mu\) using gradient descent, the loss function would be \(N-\int n(x) \;dx\), we do this to make sure the total particle number is constant
Then we calculate \(n_{\text{modified}}\) based on \(\mu\) and \(V\) using above formula
Then we update \(n\) by mixing the old \(n\) with \(n_{\text{modified}}\), that is \(n=(1-p)n+p\; n_{\text{modified}}\), the idea is similar to gradient descent, the \(p\) here can be understood as learning rate
Using the new \(n\) we can calculate \(V_{\text{int}}=wn\), then we add the interaction energy to \(V_{\text{ext}}\) as our modified \(V\), that is \(V=V_{\text{ext}}+V_{\text{int}}\)
Then we can go back to \(3\) until \(n\) converges
Simple implementation in Python
import numpy as np
import matplotlib.pyplot as plt
class SimpleDPFT:
def __init__(self,args):
self.Vext = args["Vext"]
self.maxIteration = args["maxIteration"]
self.mu = args["mu"]
self.alpha = args["alpha"]
self.N = args["N"]
self.dx = args["dx"]
self.learningRate = args["learningRate"]
self.gradientMax = args["gradientMax"]
self.w = args["w"]
self.p = args["p"]
self.nChangeMax = args["nChangeMax"]
def selfConsistentLoop(self):
self.V = self.Vext
self.mu = self.enforceN(self.mu)
self.n = self.getDensity(self.mu)
for i in range(self.maxIteration):
self.V = self.Vext + self.w * self.n
self.mu = self.enforceN(self.mu)
nOld = self.n
self.n = (1-self.p)*self.n + self.p*self.getDensity(self.mu)
if(np.sum(abs(self.n-nOld))<self.nChangeMax):
break
return self.n
def enforceN(self, mu):
# gradient descent
for i in range(self.maxIteration):
gradient = (self.cost(mu+1)-self.cost(mu))
mu = mu - self.learningRate * gradient
if(abs(gradient)<self.gradientMax):
break
return mu
def cost(self, mu):
return abs(self.N - np.sum(self.getDensity(mu)*self.dx) )
def getDensity(self,mu):
muMinusV = mu - self.V
muMinusV[ muMinusV<0 ] = 0
return np.sqrt(1/self.alpha * muMinusV)
# =================================================
# ================= Usage =========================
# =================================================
x = np.linspace(-10, 10, num=100)
dx = x[1]-x[0]
args = {
"Vext": x**2,
"maxIteration": 4000,
"mu": 1,
"alpha": np.pi**2/2,
"N": 20,
"dx": dx,
"learningRate": 1e-1,
"gradientMax": 1e-6,
"w": 5,
"p": 0.5,
"nChangeMax": 1e-6,
}
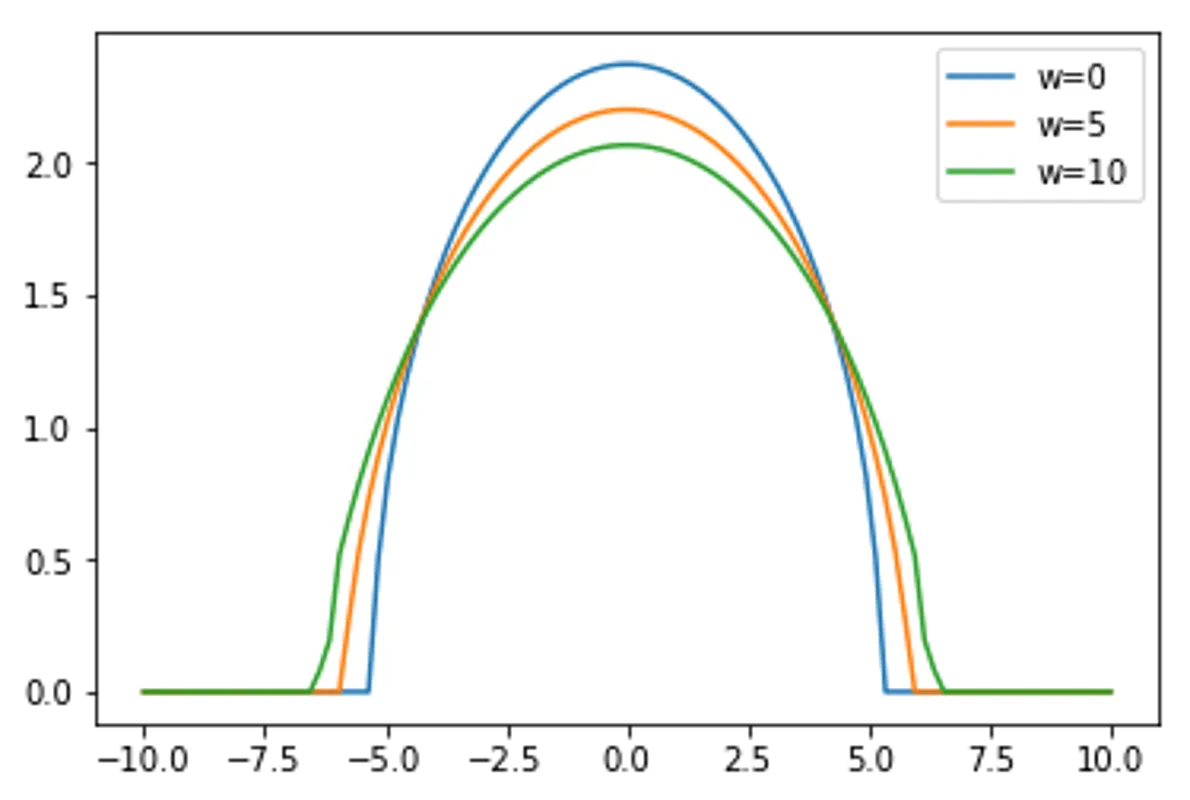
for w in range(3):
args["w"] = w*5
simpleDPFT = SimpleDPFT(args)
n = simpleDPFT.selfConsistentLoop()
print("Particle Number Check:", np.sum(n)*dx,"/ 20")
plt.plot(x,n,label="w="+str(w*5))
plt.legend()
plt.show()
Particle Number Check: 19.651993996020014 / 20
Particle Number Check: 19.524746349214976 / 20
Particle Number Check: 19.440909250694254 / 20